Ecological Archives E093-165-A2
Jay T. Lennon, Zachary T. Aanderud, B. K. Lehmkuhl, and Donald R. Schoolmaster, Jr. 2012. Mapping the niche space of soil microorganisms using taxonomy and traits. Ecology 93:1867–1879. http://dx.doi.org/10.1890/11-1745.1
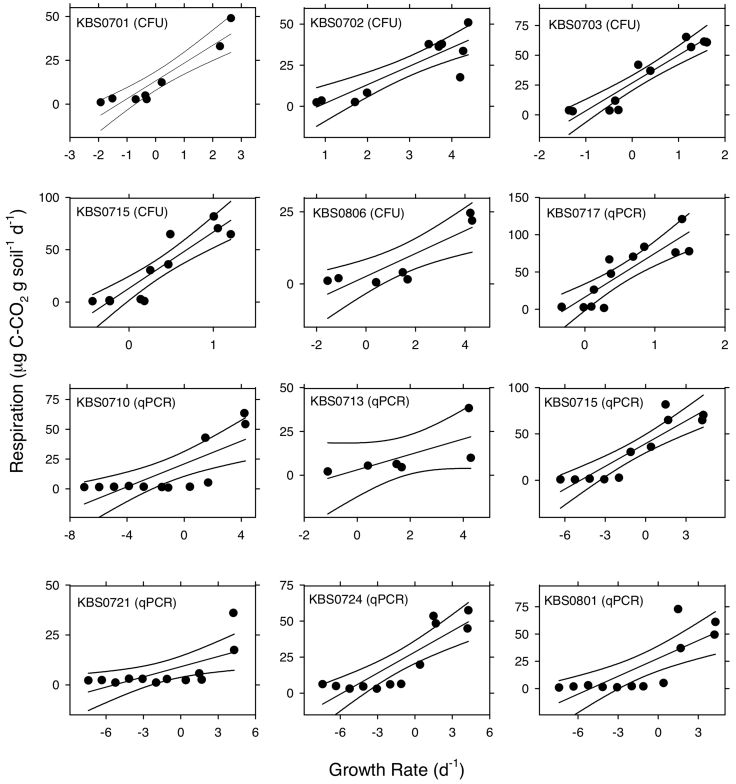
Appendix B (Fig. B1). The relationships between population growth rates and respiration rates for a subset of the bacterial strains used in this study. Bacteria were incubated in soil microcosms along a water potential gradient as described in the manuscript. Respiration rates were quantified as described in the manuscript. We measured bacterial growth rate in each microcosm as [ln(Ntf) / ln(Nt0)] / t, where Ntf is the bacterial population size at the end of an incubation, Nt0 is the bacterial population size at the beginning of the incubation, and t is the duration of the incubation. We estimated bacterial population sizes using colony forming units (CFU) and quantitative PCR (qPCR). For CFU-based estimates of bacterial population sizes, we extracted bacteria from 1 g of soil in 10 mL of 1% sodium pyrophosphate before suspensions were plated onto R2 agar. For qPCR-based estimates of bacterial population sizes, we estimated copy number of the 16S rRNA gene. Details on the qPCR approach, including DNA extraction, primers, standards, and assay conditions, can be found elsewhere (Lau and Lennon 2011). Lines in each panel represent predicted values and corresponding 95% confidence intervals from simple linear regressions. "CFU" and "qPCR" in the captions of each panel indicate the method used for quantifying bacterial population densities for growth estimates. The taxonomic identity of each strain can be referenced in Appendix A.
Literature Cited
Lau, J. A., and J. T. Lennon. 2011. Evolutionary ecology of plant-microbe interactions: soil microbial structure alters natural selection on plant traits. New Phytologist. 192:215–224