Ecological Archives A025-112-A1
Justin G. Schuetz, Gary M. Langham, Candan U. Soykan, Chad B. Wilsey, Tom Auer, and Connie C. Sanchez. 2015. Making spatial prioritizations robust to climate change uncertainties: a case study with North American birds. Ecological Applications 25:1819–1831. http://dx.doi.org/10.1890/14-1903.1
Appendix A. Additional methods and figures related to species distribution modeling approaches and sensitivity analyses.
A.1 Species distribution models
Presence/absence data were obtained from two sources: the North American Breeding Bird Survey (BBS) (Sauer et al. 2012) and the Audubon Christmas Bird Count (CBC) (Link et al. 2006). These surveys are remarkable in the breadth of spatial coverage and the use of consistent sampling methods. The BBS monitors bird populations in the United States and Canada between early May and mid-July. Survey routes are 39.4 km long with 50 stops. At each stop, participants conduct a 3-min point count and record birds seen or heard. For BBS routes, we distilled raw counts of each species over the first 30 stops (~24 km) into presence/absence data in an effort to balance the spatial scales of the BBS and CBC (see below) and to maintain a reasonable match to the resolution of the climate grids that we used for projections (10 × 10 km). The CBC began in 1900 to document early winter bird assemblages in North America and has a growing presence throughout the Western Hemisphere. CBC surveys are conducted within 24.1 km-diameter circles for one 24 h period during a four-week interval centered on December 25. For this study, all circles that fell within the boundaries of the United States and Canada were included. For every circle and count year, we distilled raw count data into presence/absence information for each species.
Historical climate data were obtained from the Canadian Forest Service (CFS) and linked to historical bird observations using a point extraction tool that provided spatially continuous estimates of climate across the landscape (McKenney et al. 2011). By using the mid-point of each CBC circle and the start-point of each BBS route as extraction points we ensured the distance between climate estimates and bird observations never exceeded 24 km. This approach seemed reasonable given the structure of the data sets, the continental scale of the study, inherent uncertainty in historical climate estimates, and the lack of systematic bias in the association of climate estimates and bird observations (even when small spatial mismatches may have occurred). We matched bird data and climate data on an annual basis, assuming that climate variables from the year leading up to each survey would best inform our understanding of occurrence data. We used 17 bioclimatic variables to characterize conditions in each year (Distler et al. in press). Bioclimatic variables are thought to represent biologically meaningful combinations of monthly climate variables because they aggregate climate information in ways known to drive biological processes (Phillips et al. 2006). Bioclimatic variables can be highly correlated, however, which raises concerns about the ability of modeling algorithms to identify causal relationships in data sets and to generate accurate predictions (Dormann et al. 2013). A variety of approaches to clustering or removing predictors can be employed in an effort to reduce collinearity but none of them guarantee better inference or prediction (Dormann et al. 2013). Ultimately, iterative assessment of correlated variables through observation and experiment is needed to establish which are the true drivers of biological processes. We chose not to remove variables from our data set because we had little or no a priori justification for choosing one correlated variable over another.
Species distribution models were developed using boosted regression trees (Ridgeway 2010). We built separate distribution models for breeding and non-breeding seasons using BBS and CBC data, respectively. Our analyses of BBS and CBC data were similar in approach with small adjustments to account for differences in data sets and survey protocols. In addition to bioclimatic variables, for BBS analyses we used Julian date to account for variation in timing of surveys across the summer season. We used 25081 records collected along 3718 routes from 2000–2009 to train our models and 41959 records collected from 1980–1999 to test the predictive performance of our models. This approach allowed us to take advantage of increased geographic sampling in recent years to build models and availability of abundant historical data for assessing the predictive ability of our models outside the current time period and climate space. We had sufficient data to construct models for 475 North American species during the breeding season. For CBC analyses, we included the number of survey hours invested in each CBC circle as a predictor to account for uneven observer effort across circles, in addition to bioclimatic variables. The number of participating individuals and the duration of counts vary among CBC circles and through time, thus the number of party-hours has often been used as a covariate to account for this variation in analyses based on CBC data (Dunn et al. 2005, Link and Niven 2006). We used 19272 records collected at 2278 circles from 2000–2009 to train our models and 30630 records collected from 1980–1999 to assess the predictive ability of our models. We had sufficient data to construct models for 503 North American species during the non-breeding season.
We generated current species distributions by projecting modeled relationships across gridded annual bioclimatic data averaged from 1999–2008, the same period used to construct the models. A spatial layer containing the median survey day was included as a constant in projections of BBS models and a layer containing the median number of survey hours was included as a constant in projections of CBC models. To characterize future climates and establish a spatial context for projections, we added future climate anomaly grids to baseline climate data in several processing steps. First, we obtained spatially downscaled (5-min resolution) WorldClim climate grids for 13 combinations of general circulation models (GCMs) and emissions scenarios (Distler et al. in press) and summarized across three future time periods (2010–2039, 2040–2069, 2070–2099; hereafter 2020s, 2050s, 2080s). We then subtracted contemporary WorldClim grids for monthly minimum temperature, maximum temperature, and precipitation from the future grids to isolate predicted climate anomalies from baseline values. Finally, we added these monthly anomaly grids to CFS mean climate grids for the base period (1971–2000), ensuring correspondence between the historical climate used to build the models and simulated future climate (Fischlin et al. 2007). We then transformed projected temperature and precipitation data into bioclimatic variables using DIVA software and the raster package (Hijmans and van Etten 2011) in the statistical software R.
Species distribution models were projected into each of 39 future climate surfaces (i.e., 13 combinations of emissions scenarios and GCMs in each of 3 future time periods). Because we were fundamentally interested in understanding responses of species to climate change across scenarios and time, we ensembled projections for multiple GCMs within each scenario and time period (consensus forecasting sensu Araújo and New 2007). This process resulted in 9 future prediction grids for each species, one for each emissions scenario (SRES B2, A1B, A2) in each time period (2020, 2050, 2080) that described climatic suitability of the United States and Canada on a continuous scale from 0 (unsuitable) to 1 (highly suitable).
We performed additional analyses to assess how sensitive our findings were to the number and identity of taxa included in the prioritization process. The following sets of species were analyzed according to the same methods described in the text (see Supplement for species lists).
Season |
Number of species in prioritization |
Number of samples |
winter |
1 |
503 |
winter |
5 |
40 |
winter |
10 |
20 |
winter |
50 |
20 |
winter |
149 |
10 |
winter |
503 |
1 |
Season |
Number of species in prioritization |
Number of samples |
summer |
1 |
475 |
summer |
5 |
40 |
summer |
10 |
20 |
summer |
50 |
20 |
summer |
233 |
10 |
summer |
475 |
1 |
Single-species prioritizations were generated for every species for which we had a species distribution model (i.e., 503 winter distributions and 475 summer distributions). Species lists for all other samples were generated using random draws (with replacement) from the complete set of taxa available for each season (see Supplement for species lists).
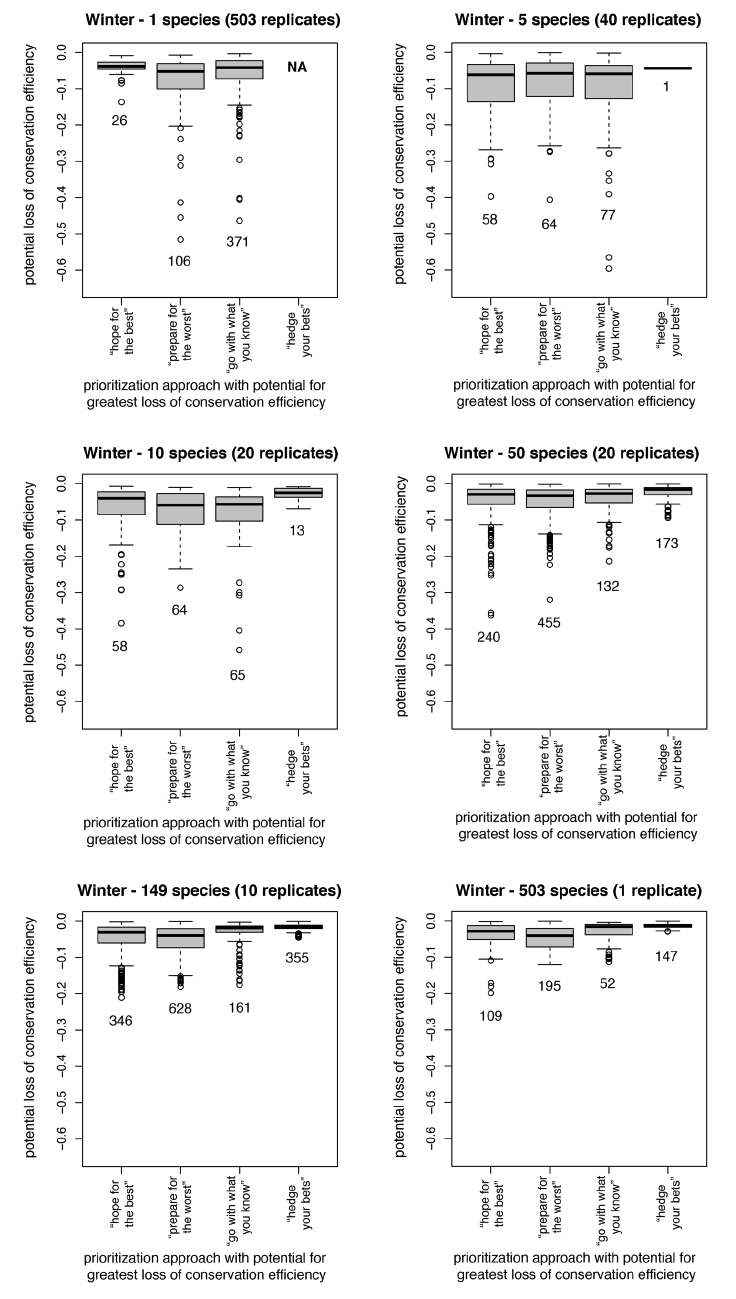
Fig. A1. Sensitivity analyses describing conservation inefficiency for individual species when biological assumptions and biological responses are mismatched during winter. For each species, we identified the prioritization approach with the greatest potential for misrepresenting its conservation needs (see Supplement for species list). Loss values reflect the decline in conservation efficiency for an individual species between a prioritization in which biological assumptions and biological responses match and a prioritization in which the mismatch between assumptions and responses leads to the greatest loss of efficiency. Descriptions above each plot indicate the size of each prioritization (i.e., how many species were included) and the number of replicates. Values below each box within a boxplot indicate the number of species for which that prioritization had the potential to be most inefficient.
Fig. A2. Sensitivity analyses describing conservation inefficiency for individual species when biological assumptions and biological responses are mismatched during summer. For each species, we identified the prioritization approach with the greatest potential for misrepresenting its conservation needs (see Supplement for species list). Loss values reflect the decline in conservation efficiency for an individual species between a prioritization in which biological assumptions and biological responses match and a prioritization in which the mismatch between assumptions and responses leads to the greatest loss of efficiency. Descriptions above each plot indicate the size of each prioritization (i.e., how many species were included) and the number of replicates. Values below each box within a boxplot indicate the number of species for which that prioritization had the potential to be most inefficient.
Literature cited
Araújo, M. B., and M. New. 2007. Ensemble forecasting of species distributions. Trends Ecol. Evol., 22:42–47.
Distler, T., J. G. Schuetz, J. Velásquez-Tibatá, and G. M. Langham. In Press. Stacked species distribution models and macroecological models provide congruent projections of avian species richness under climate change. Journal of Biogeography.
Dormann, C. F., J. Elith, S. Bacher, C. Buchmann, G. Carl, G. Carré et al. 2013. Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography, 36:27–46.
Dunn, E. H., C. M. Francis, P. J. Blancher, S. Roney Drennan, M. A. Howe, D. Lepage, et al. 2005. Enhancing the scientific value of the Christmas Bird Count. Auk, 122:338–346.
Fischlin, A., et al. in IPCC Climate Change 2007: Impacts, Adaptation and Vulnerability, M. L. Parry, O. F. Canziani, J. P. Palutikof, P. J. van der Linden, and C. E. Hanson, Editors. Cambridge University Press, pp. 211–272.
Hijmans, R. J., and J. van Etten. 2011. Raster: Geographic analysis and modeling with raster data. R package version 1, r948.
Phillips, S. J., R. P. Anderson, and R. E. Schapire. 2006. Maximum entropy modeling of species geographic distributions. Ecol. Model. 190:231–259.
Sauer, J., J. E. Hines, J. E. Fallon, K. L. Pardieck, D. J. Ziolkowski, and W. A. Link. 2012. The North American Breeding Bird Survey, results and analysis 1966–2009. USGS, Laurel, Maryland. Version 3.23.